Difference between revisions of "Cryotherapy"
| Line 3: | Line 3: | ||
== Additional information == | == Additional information == | ||
| − | |||
| − | |||
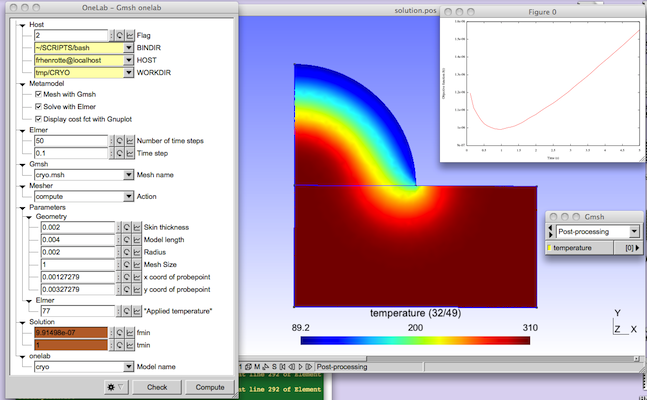
| − | {{metamodelfooter| | + | The physical background is the cryogenic treatment of warts by application of a cryogenic fluid. The idea is to maximize the destruction of wart tissue cells while minimizing damages to healthy skin tissue. A damage function depending on temperature distribution and exposure time is built to represent this trade-off. The purpose of the modeling is to determine the application time that minimizes the damage function. |
| + | Geometrical and modeling parameters can be interactively modified in the ONELAB window. By clicking on the "Run" button, the thermal solver of Elmer is invoked. After execution, a plot of the damage function vs. time is displayed, and the computed optimum application time tmin is highlighted in the ONELAB window. | ||
| + | |||
| + | Launch Gmsh and open ELMERMODELS/CRYO/cryo.py from the "File" menu. | ||
| + | |||
| + | {{metamodelfooter|cryotherapy}} | ||
Revision as of 10:44, 26 May 2014
|
2D tool for cryotherapy.
|
|
|---|
|
Download model archive (cryotherapy.zip) |
Additional information
The physical background is the cryogenic treatment of warts by application of a cryogenic fluid. The idea is to maximize the destruction of wart tissue cells while minimizing damages to healthy skin tissue. A damage function depending on temperature distribution and exposure time is built to represent this trade-off. The purpose of the modeling is to determine the application time that minimizes the damage function. Geometrical and modeling parameters can be interactively modified in the ONELAB window. By clicking on the "Run" button, the thermal solver of Elmer is invoked. After execution, a plot of the damage function vs. time is displayed, and the computed optimum application time tmin is highlighted in the ONELAB window.
Launch Gmsh and open ELMERMODELS/CRYO/cryo.py from the "File" menu.
|
Model developed by F. Henrotte.
|

