Elmer
Elmer is an open source (GPL) computational tool for multi-physics problems. It is developed by CSC in collaboration with Finnish universities, research laboratories and industry.
Two complete metamodels using Elmer are available on the read-only svn distribution of gmsh, which can be downloaded from http://www.geuz.org/gmsh.
You also need a working recent (nightly-build) version of gmsh to be executable on your system.
First take a look at this tutorial to see how ONELAB can be installed.
LASER
The metamodel "LASER" is launched as follows:
- cd $GMSH_DIR/projects/onelab/METAMODELS/LASER (the variable GMSH_DIR refers to the home directory of your Gmsh installation)
- gmsh lab_peau.geo -s
The shell variable GMSH_DIR refers to the home directory of your Gmsh installation, see Tutorial. The "-s" option amounts to click on "Tool/Onelab" in the Gmsh menu.
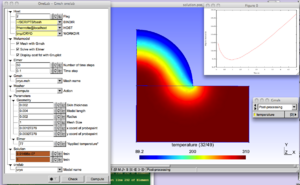
The physical background of this model is the laser stimulation of skin in order to measure the density of nociceptive receptors. For a correct interpretation of the experimental data, an accurate nowledge of the temperature distribution in time and across the skin is needed. The metamodel allows selecting various models of stimulation (imposed flux, imposed temperature and controlled temperature) and various laser types (Gaussian or flat-top). Each simulation generates a Matlab or Gnuplot graphical output that is directly interpretable by clinicians. Check in the "lab_peau.ol" file how the switch between Matlab and Gnuplot is done, and modify the file if necessary according to your system settings.
CRYO
The cryo-engineering metamodel "CRYO" is launched as follows:
- cd $GMSH_DIR/projects/onelab/METAMODELS/CRYO
- gmsh cryo.geo -s
The shell variable GMSH_DIR refers to the home directory of your Gmsh installation, see Tutorial. The "-s" option amounts to click on "Tool/Onelab" in the Gmsh menu.
The physical background is the cryogenic treatment of warts by application of a cryogenic fluid. The idea is to maximize the destruction of wart tissue cells while minimizing damages done to healthy skin tissue. An damage function depending on temperature distribution and time is built to represent this trade-off and the purpose of the modeling is to determine the application time that minimizes this damage function.
Various geometrical and modeling parameters can be interactively modified in the ONELAB window.
After execution, a plot of the damage function displayed and the optimum application time tmoin is uploaded to the ONELAB window.
This metamodel illustrates also the possibility of running a client on a remote host. After selecting the option "remote" of the parameter "host" and clicking on "Check", empty boxes will appear to be filled in with information about a remote host (user@remote), a remote working directory, and a remote bin directory where the executable of the remote client can be found.
These pieces of information can be entered directly in the interactive box. They will be stored on the server and be available for the duration of the ONELAB session.
Alternatively, if entered directly in the file cryo.ol in the definition of the variables HOST, REMDIR and ELMERDIR, they will be available for all ONELAB sessions.
As long as HOST and REMDIR are empty, the client is solved on the local machine, whatever the value of the flag host.