Elmer
Elmer is an open source (GPL) computational tool for multi-physics problems. It is developed by CSC in collaboration with Finnish universities, research laboratories and industry.
Two complete metamodels using Elmer are available on the read-only svn distribution of gmsh, which can be downloaded from geuz.org/gmsh.
You also need a working recent (nightly-build) version of gmsh to be executable on your system.
First take a look at [tutorial] to see how ONELAB can be installed.
LASER
The cryo-engineering metamodel "LASER" is launched as follows:
- export GMSH_DIR=/path/to/your/gmsh/directory
- cd $GMSH_DIR/projects/onelab/METAMODELS/CRYO
- gmsh circle.geo -s
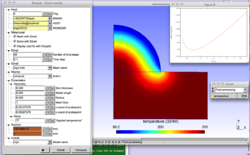
The physical background is the laser stimulation of skin in order to measure the density [[1]] receptors. For a correct interpretation of the experimental data, an accurate nowledge of the temperature distribution in time and space is needed. The metamodel offers various kind of stimulation (imposed flux, imposed temperature and controlled temperature) and various lasers (Gaussian or flat-top). Each simulation generates a Matlab or Gnuplot graphical output that is directly interpretable by clinicians. Check in the "lab_peau.ol" file how the switch between Matlab and Gnuplot is done, and modify the file if necessary according to your system settings.
CRYO
The cryo-engineering metamodel "CRYO" is launched as follows:
- export GMSH_DIR=/path/to/your/gmsh/directory
- cd $GMSH_DIR/projects/onelab/METAMODELS/CRYO
- gmsh circle.geo -s
The physical background is the cryogenic treatment of warts. The wart tissue must be