Elmer
Introduction
Elmer is an open source (GPL) computational tool for multi-physics problems. It is developed by CSC in collaboration with Finnish universities, research laboratories and industry. To test ONELAB models working with Elmer, you first need to install the generic ONELAB virtual machine on your system by following these instructions. When done, you can log in into the virtual machine (username: "olvm", passwd: olvm) and proceed with the installation of the Elmer software, and the download of benchmark ONELAB models. The instructions to do so are given in the next section.
Installation
The Virtual machine is configured so that Gmsh and Elmer can be installed straightforwardly by issuing simple commands in a terminal. Proceed as follows.
- Step 1
- Download installation scripts
- Open a terminal by clicking on the "terminal" icon in the launcher panel and issue the command
install_scripts.sh
Then, the command
install_gmsh.sh
will download the nightly-build of Gmsh from http://geuz.org/gmsh, install it, and place a Gmsh icon on the Desktop.
When done, issue the command
install_elmer.sh
which will download the source code of Elmer and compile it on the virtual machine. This may take several minutes.
Now, you have a woking installation of Gmsh and Elmer on the virtual machine. A number of Elmer model examples can be downloaded by issuing the command
getElmerModels.sh
After all, you have a directory "ELMERMODELS" in your home directory with a number of Elmer models, which are now presented with more details.
ONELAB models
CRYO
Launch Gmsh and open ELMERMODELS/CRYO/cryo.py from the "File" menu.
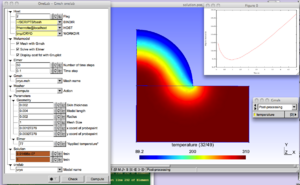
The physical background is the cryogenic treatment of warts by application of a cryogenic fluid. The idea is to maximize the destruction of wart tissue cells while minimizing damages to healthy skin tissue. A damage function depending on temperature distribution and exposure time is built to represent this trade-off. The purpose of the modeling is to determine the application time that minimizes the damage function.
Geometrical and modeling parameters can be interactively modified in the ONELAB window. By clicking on the "Run" button, the thermal solver of Elmer is invoked. After execution, a plot of the damage function vs. time is displayed, and the computed optimum application time tmin is highlighted in the ONELAB window.
BEAM
This model is the didactical analysis of a clamped beam (static 3D elasticity). The dimensions of the beam and the material parameter can be modified interactively in the ONELAB window, as well as a number of modeling parameters. Diagrams of the internal moments can be generated by checking the box Compute MT diagrams.
Launch Gmsh and open ELMERMODELS/BEAM/beam.py from the "File" menu.
PATCH
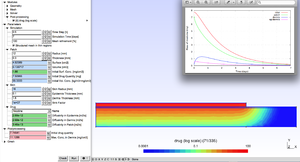
This metamodel is toolkit for drug patch conception.
The user can determine the geometry of the patch and initial drug concentrations.
A number of drugs are pre-tabulated with their diffusivities.
Elmer solves a convection-diffusion equation and post processing tools display the rate at which drug is transmitted to the skin.
LASER
Download and inflate the archive LASER.zip in a work directory. Right-click on the icon laser.ol and open the file with gmsh. Alternatively, start gmsh and click File > Open > laser.ol from the menu.
The physical background of this model is the laser stimulation of skin in order to measure the density of nociceptive receptors. For a correct interpretation of the experimental data, an accurate knowledge of the temperature distribution in time and across the skin is needed. The metamodel allows selecting various laser types (Gaussian, flat-top) and various stimulus characteristics (imposed flux or controlled temperature). Each simulation generates a graphical result file plot.pdf that is directly interpretable by clinicians.